
Probabilistic Machine Learning
Research of the Probabilistic Machine Learning Group
We develop new methods for probabilistic modeling, Bayesian inference and machine learning. Our current focuses are:
Amortized Experimental Design, Optimization and Simulation-based Inference
Bayesian experimental design (BED), Bayesian optimization (BO), and simulation-based inference (SBI) are driving forces in science and engineering as principled frameworks for collecting high-quality data and parameter estimation, respectively. A fundamental challenge with these frameworks is computational scalability due to the need for repeated inference. BED and BO involve repeating a lengthy process for estimating and optimizing the information gain or the acquisition function, respectively, for each new query, while in SBI, running costly posterior sampling methods (e.g., MCMC) for every new observation is a bottleneck. We tackle these challenges by developing methods that are “amortized”, i.e., methods that pay an upfront cost of training a deep learning model to circumvent a recurring computationally expensive step. Amortized experimental design, optimization, and SBI methods therefore accelerate scientific discovery and data analysis in many fields of science and engineering, making Bayesian inference scalable for complex problems.
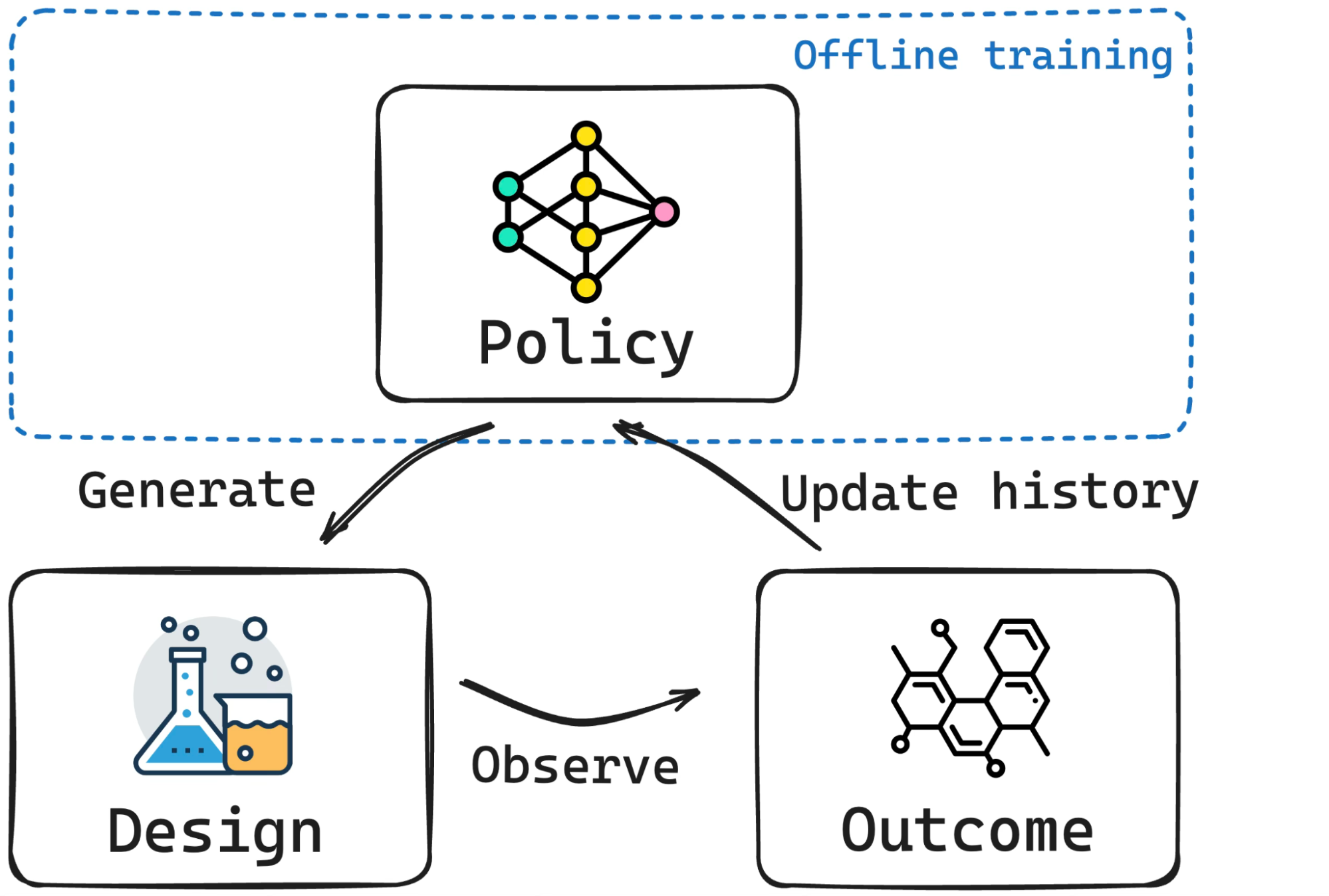
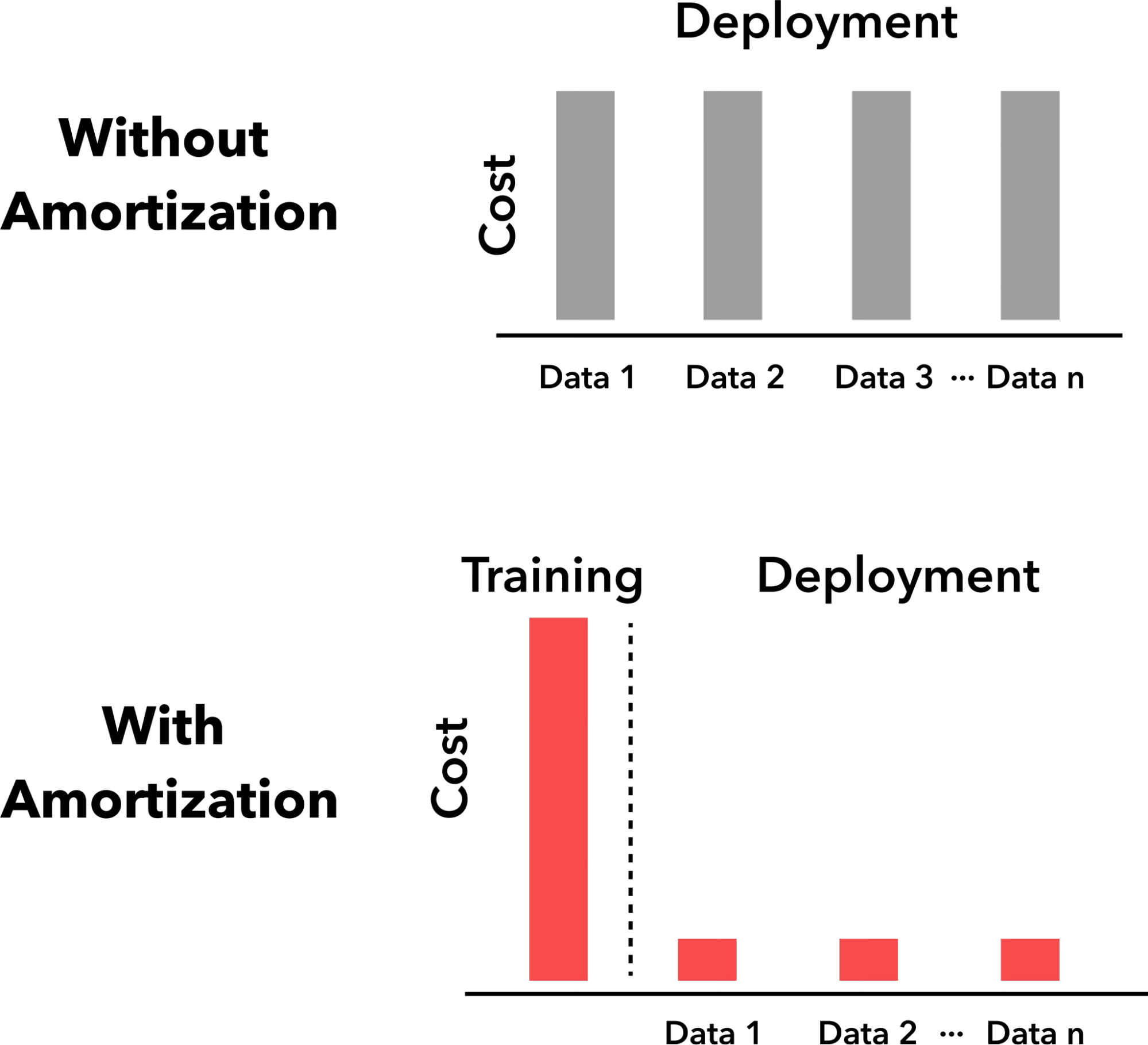
Reference
- Huang, D., Wen, X., Bharti, A., Kaski, S. & Acerbi, L. (2025), ALINE: Joint Amortization for Bayesian Inference and Active Data Acquisition, Advances in Neural Information Processing Systems (NeurIPS), Spotlight.[Link.]
- Bharti, A., Huang, D., Kaski, S. & Briol, F.-X. (2025), Cost-aware simulation-based inference, Proceedings of the International Conference on Artificial Intelligence and Statistics (AISTATS).[Link.]
- Chang, P., Loka, N., Huang, D., Remes, U., Kaski, S. & Acerbi, L. (2025), Amortized Probabilistic Conditioning for Optimization, Simulation and Inference, Proceedings of the International Conference on Artificial Intelligence and Statistics (AISTATS). [Link.]
- Zhang, X., Huang, D., Kaski, S. & Martinelli, J. (2025), PABBO: Preferential Amortized Black-Box Optimization, International Conference on Learning Representations (ICLR), Spotlight.[Link.]
- Huang, D., Guo, Y., Acerbi, L. & Kaski, S. (2024), Amortized Bayesian Experimental Design for Decision-Making, Advances in Neural Information Processing Systems (NeurIPS). [Link.]
- Sinaga, M.A., Martinelli, J. and Kaski, S. (2025), Robust and computation-aware Gaussian processes. Proceedings of the 39th Annual Conference on Neural Information Processing Systems (NeurIPS 2025).[Link.]
Generative Modeling
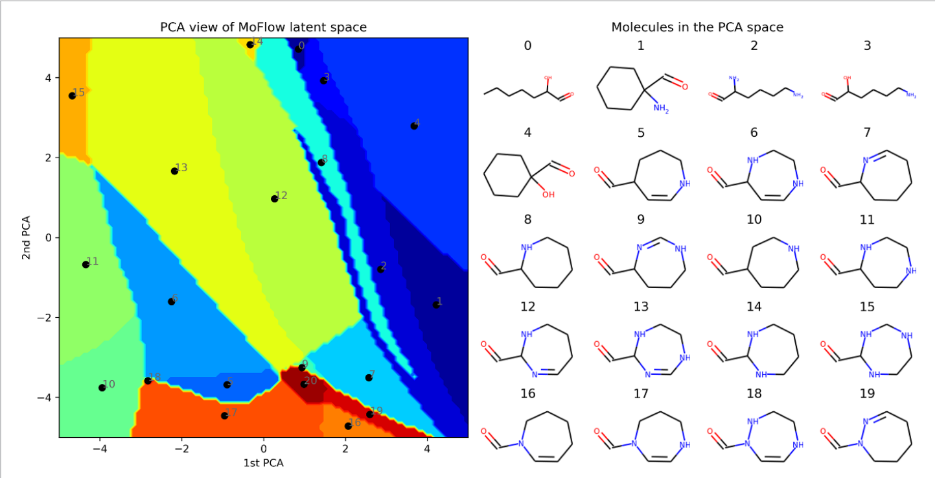
Generative models offer powerful tools to represent complex data distributions, with recent advances, such as flow-matching and diffusion models, providing expressive, high-dimensional priors. Together, these approaches form two key pillars for scientific applications. In generative modeling, our research focuses on improving model reliability when data is scarce or when prior expert knowledge is valuable. We develop methods for reusing and adapting pretrained generative models, integrating human-in-the-loop feedback, and reducing epistemic uncertainty in generated outputs. Our applications include molecular and image generation as well as natural language-based scientific interaction.
Reference
- Alakuijala, M., Gao, Y., Ananov, G., Kaski, S., Marttinen, P., Ilin, A. and Valpola, H. (2025) ‘Memento no more: Coaching AI agents to master multiple tasks via hints internalization’, arXiv preprint, arXiv:2502.01562. [Link].
- Alakuijala, M., McLean, R., Woungang, I., Farsad, N., Kaski, S., Marttinen, P. and Yuan, K. (2025) ‘Video-Language Critic: Transferable reward functions for language-conditioned robotics’, Transactions on Machine Learning Research, 2025, pp. 1-22.[Link].
- Jain, A., Heinonen, M., Käsnänen, H., Sipilä, J. and Kaski, S., 2025. Multi-target property prediction and optimization using latent spaces of generative model. Machine Learning: Science and Technology, 6(2), p.025042. [Link].
- Garipov, T., De Peuter, S., Yang, G., Garg, V., Kaski, S. and Jaakkola, T., 2023. Compositional sculpting of iterative generative processes. Advances in neural information processing systems, 36, pp.12665-12702. [Link].
ATOM (Artificial agents with Theory Of Mind) and Human-In-The-Loop
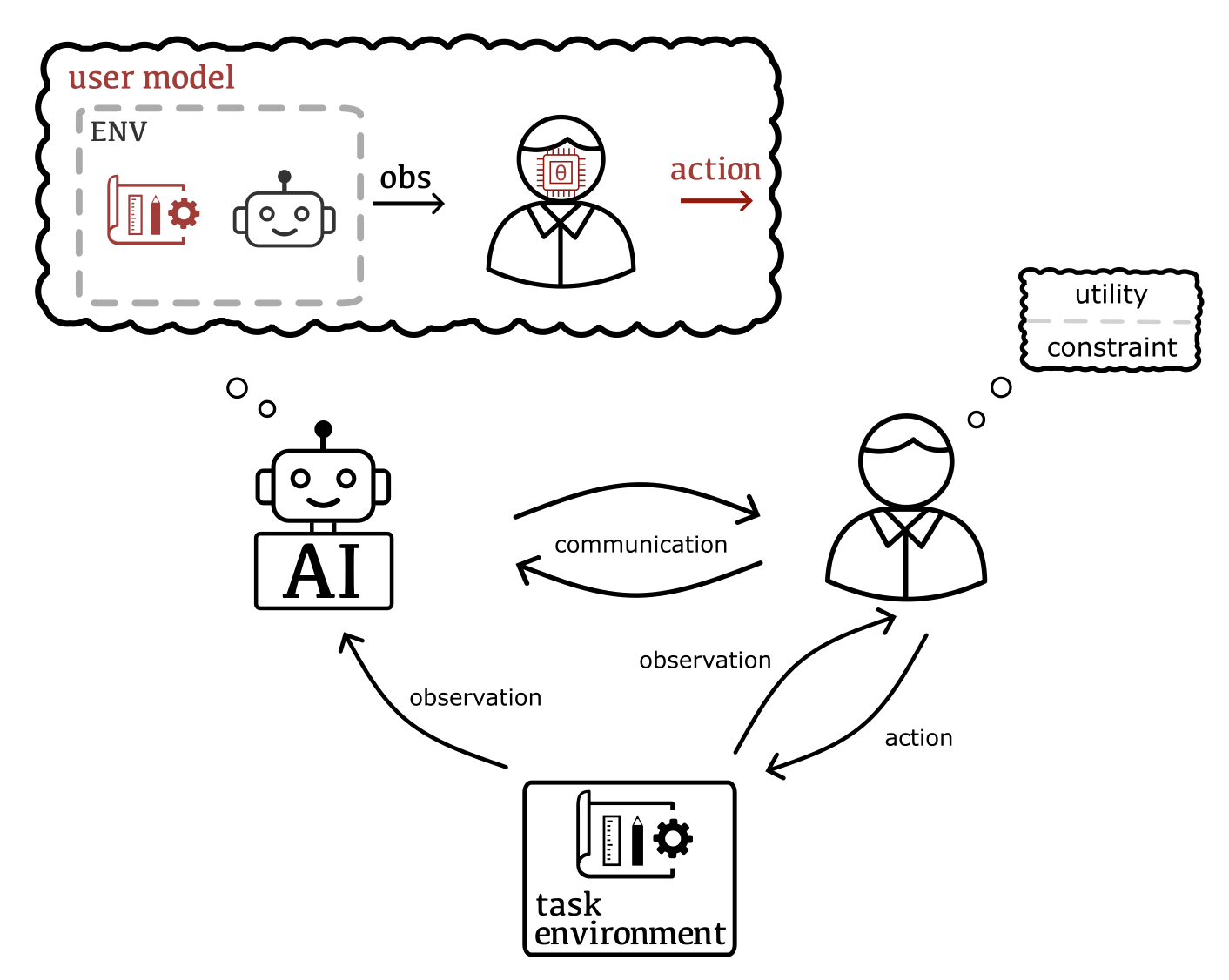
Critical problems tackled by machine learning involve human users in the loop. Typically, their solutions do not account for the users as individuals and hence fail to adapt to the tacit and evolving needs of their users. Addressing this caveat requires reasoning about the implications of human behavior and, reversely, taking the user's understanding of their own process (i.e., the machine learning system) into account. This includes e.g. inferring tacit and changing goals and knowledge, and modeling how humans strategically adapt to the system based on how they interpret its behavior.
Our research aims to move away from systems in which users are treated as passive sources of data, towards expecting users to be strategic and collaborative. Our methods combine Bayesian inference and decision making, multi-agent reasoning, and ideas in cognitive science such as theory of mind and computational rationality. Output of this research are scalable posterior approximation methods, for example inference over the user, novel frameworks for human-AI decision making, and user models for realistic and sophisticated settings, such that our AI can truly collaborate with humans.
Keywords: human-AI collaboration, Bayesian inference, sequential (multi-agent) decision making, user modeling, theory-of-mind.
Reference
- De Peuter, Sebastiaan, Shibei Zhu, Yujia Guo, Andrew Howes, and Samuel Kaski. Preference learning of latent decision utilities with a human-like model of preferential choice. Advances in Neural Information Processing Systems 37 (2024): 123608-123636. [Link].
- De Peuter, Sebastiaan, and Samuel Kaski. Zero-shot assistance in sequential decision problems. In Proceedings of the AAAI Conference on Artificial Intelligence, vol. 37, no. 10, pp. 11551-11559. 2023. [Link].
- Mustafa Mert Çelikok, Pierre-Alexandre Murena, Samuel Kaski (2023). Modeling needs user modeling. Frontiers in Artificial Intelligence, 6:1097891. [Link].
- Hämäläinen, Alex, Mustafa Mert Çelikok, and Samuel Kaski. Differentiable user models. In Uncertainty in Artificial Intelligence, pp. 798-808. PMLR, 2023. Best paper award in NeurIPS HILL Workshop 2022 for a preliminary version. [Link.]
- De Peuter, Sebastiaan, Antti Oulasvirta, and Samuel Kaski. Toward AI assistants that let designers design. Ai Magazine 44, no. 1 (2023): 85-96.[Link.]
Robustness to Distribution Shifts in Deployment
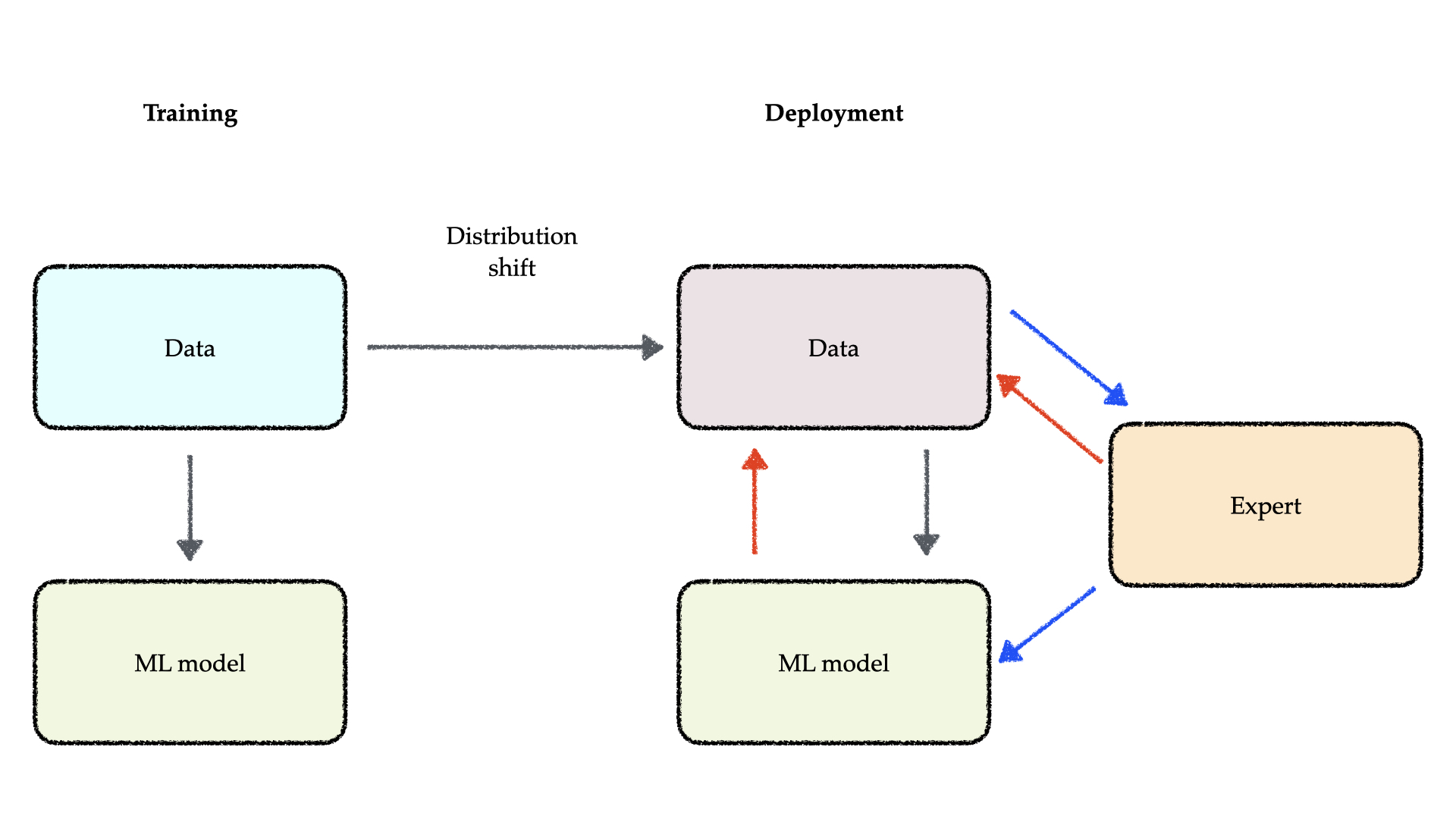
The immense promise of machine learning is often challenged by its practical deployment, which faces a critical hurdle: distribution shifts. A core assumption in machine learning is that future data will follow the same statistical distribution as the training data. However, this assumption is frequently violated in the real world, since the underlying data-generating processes can shift over time or differ across contexts. Such shifts typically cause a sharp decline in model performance, due to issues like model misspecification, unrepresentative data sampling, or spurious correlations. Our work focuses on developing principled methods for building machine learning systems that are robust to unexpected confounders, making them apt for deployment in critical applications such as personalized medical treatment recommendation and drug discovery.
To this end, our research addresses the core technical hurdles that prevent robust learning. We have developed techniques to improve the transfer of knowledge between different but related tasks, such as mitigating the effects of prior misspecification when adapting probabilistic models to new data (Sloman, et al., 2025). Furthermore, to ensure models generalize effectively across diverse environments and avoid the pitfalls of negative transfer, we have designed novel meta-learning approaches that capture the similarity of underlying causal mechanisms across datasets (Wharrie, et al., 2024).
Reference
- Sloman, S., J., Martinelli, J., and Kaski, S. (2025), Proxy-informed Bayesian transfer learning with unknown sources, The 41st Conference on Uncertainty in Artificial Intelligence. [Link].
- Wharrie, S., Eick, L., Mäkinen, L., Ganna, A., Kaski, S., and FinnGen (2024), Bayesian Meta-Learning for Improving Generalizability of Health Prediction Models With Similar Causal Mechanisms. Preprint. [Link].
- Huang, D., Bharti, A., Souza, A., Acerbi, L., & Kaski, S. (2023). Learning robust statistics for simulation-based inference under model misspecification. Advances in Neural Information Processing Systems. [Link].
Privacy-preserving and Secure AI
Machine learning models are capable of unintentionally capturing sensitive information underlying the training data, leading to the disclosure of sensitive personal information. We combine differential privacy (DP), the prevailing formalism for anonymity, with Bayesian inference to develop powerful techniques for probabilistic modeling with rigorous privacy guarantees. We study the theoretical foundations and provide practical implementations for modern statistical inference under differential privacy, such as the software package Twinify, for creating and releasing anonymised synthetic twins of sensitive data sets. We also work on making DP easier to apply to new problems, supporting researchers in identifying optimal models and training configurations under DP.
Reference
- Yang, Y., Rehn, A., Katt, S., Honkela, A. and Kaski, S. (2025). An Interactive Framework for Finding the Optimal Trade-off in Differential Privacy. [Link].
- Sonee, A., Harikumar, H., Hämäläinen, A., Prediger, L. and Kaski, S. (2025). Privacy-preserving neural processes for probabilistic user modeling. UAI. [Link].
- Prediger, L., Jälkö, J., Honkela, A. and Kaski, S. (2024). Collaborative learning from distributed data with differentially private synthetic data. BMC Medical Informatics and Decision Making, 24(1), 167. [Link].
- Jälkö, J., Prediger, L., Honkela, A. and Kaski, S. (2023). DPVIm: Differentially Private Variational Inference Improved. Transactions on Machine Learning Research. [Link].
- Jälkö, J., Lagerspetz, E., Haukka, J., Tarkoma, S., Honkela, A. and Kaski, S. (2021). Privacy-preserving data sharing via probabilistic modelling. Patterns, 2:100271, 2021. [Link].
AI For Research (e.g. BioDesign, Neuroscience, Health, Drug Design, Economics)
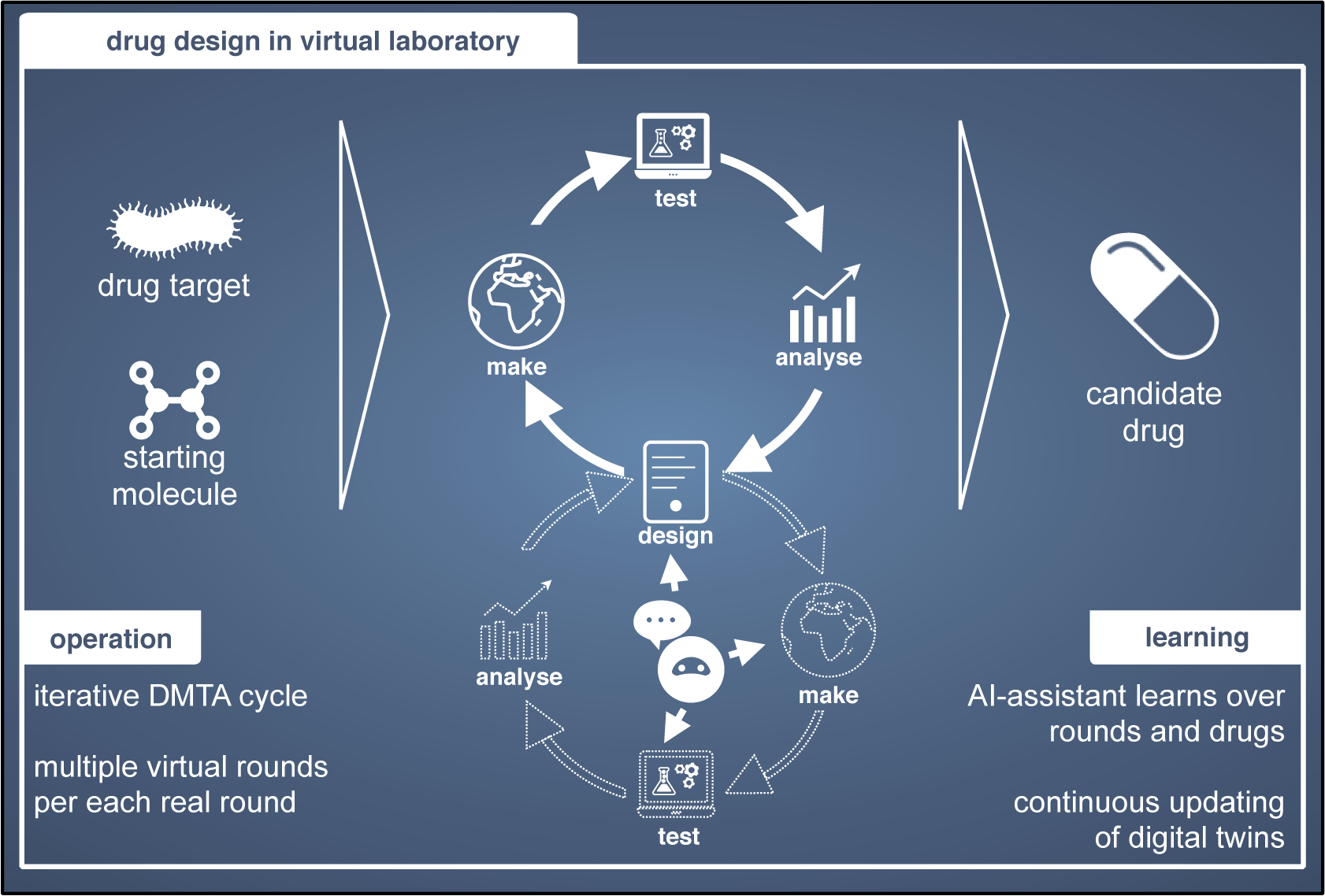
While applying AI to individual scientific domains has produced techniques such as Physics-Informed Machine Learning, we focus on AI For Research, a paradigm that enhances the research workflow itself, by optimizing Design-Build-Test-Learn (DBTL) cycles, experimental design and data acquisition, and human-in-the-loop discovery.
We work closely with researchers from other fields and industry partners to pursue breakthroughs in challenging real-world applications.
Methodologically, many of our projects center on sequential decision-making problems, spanning
healthcare, synthetic biology, drug design, cognitive science, materials science, robotics, and autonomous systems. Aligned with the FCAI Virtual Labs initiative, our recent efforts mainly split into the following strands:
Reference
- Masood, M.A., Kaski, S. & Cui, T. Molecular property prediction using pretrained-BERT and Bayesian active learning: a data-efficient approach to drug design. J Cheminform 17, 58 (2025). [Link].
- Nahal, Y., Menke, J., Martinelli, J. et al. Human-in-the-loop active learning for goal-oriented molecule generation. J Cheminform 16, 138 (2024). [Link].
- Klami A, Damoulas T, Engkvist O, Rinke P, Kaski S. Virtual laboratories: transforming research with AI. Data-Centric Engineering (2024) [Link].
- Sophie Wharrie, Zhiyu Yang, Andrea Ganna, Samuel Kaski (2023). Characterizing personalized effects of family information on disease risk using graph representation learning. In Machine Learning for Healthcare Conference (pp. 824-845). PMLR. [Link].
- Eemeli Leppäaho, Hanna Renvall, Elina Salmela, Juha Kere, Riitta Salmelin, Samuel Kaski (2019). Discovering heritable modes of MEG spectral power. Human Brain Mapping, 40(5):1391-1402. [Link].