
Probabilistic Machine Learning
GPstuff - regression demo
Gaussian Process regression demo, demo_regression1, R
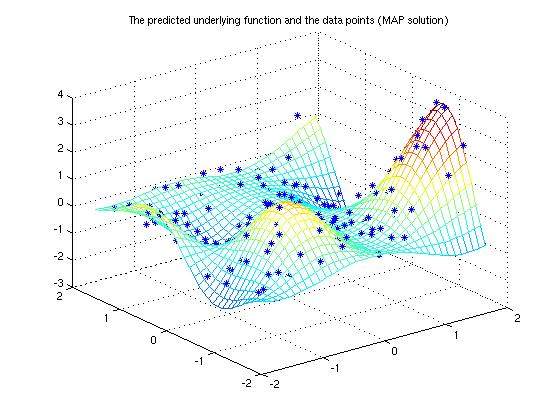
The demonstration program solves a simple 2-dimensional regression problem using Gaussian process. The data in the demonstration has been used in the work by Christopher J. Paciorek and Mark J. Schervish (2004). The training data has an unknown Gaussian noise and can be seen in Figure 1. We conduct the inference with Gaussian process using squared exponential covariance function.
Description
The regression problem consist of a data with two input variables and one output variable with Gaussian noise. The observations y are assumed to satisfy
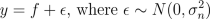
and f is an underlying function, which we are interested in. We place a zero mean Gaussian process prior for f, which implies that at the observed input locations latent values have prior
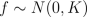
where K is the covariance matrix, whose elements are given as
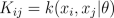
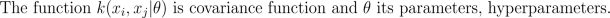
Since both likelihood and prior are Gaussian, we obtain a Gaussian marginal likelihood
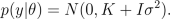
By placing a hyperprior for hyperparameters, p(th), we can find the maximum a posterior (MAP) estimate for them by maximizing
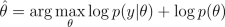
If we want to find an approximation for the posterior of the hyperparameters, we can sample them using Markov chain Monte Carlo (MCMC) methods.
After finding MAP estimate or posterior samples of hyperparameters, we can use them to make predictions for f:
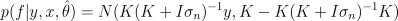
Pieces of code
Initialize GPstuff and load the data:
require(RcppOctave)
o_source(text="
P = genpath('/path/to/gpstuff/);'
addpath(P);
")
o_source(text="
S = which('demo_regression1');
L = strrep(S,'demo_regression1.m','demodata/dat.1');
data=load(L);
x = [data(:,1) data(:,2)];
y = data(:,3);
[n, nin] = size(x);
")
The model is constructed as follows:
# Create likelihood
o_source(text="
lik = lik_gaussian('sigma2', 0.2^2);
")
# Create covariance function
o_source(text="
gpcf = gpcf_sexp('lengthScale', [1.1 1.2], 'magnSigma2', 0.2^2)
")
# Set some priors
o_source(text="
pn = prior_logunif();
lik = lik_gaussian(lik,'sigma2_prior', pn);
pl = prior_unif();
pm = prior_sqrtunif();
gpcf = gpcf_sexp(gpcf, 'lengthScale_prior', pl, 'magnSigma2_prior', pm);
")
# Create GP
o_source(text="
gp = gp_set('lik', lik, 'cf', gpcf);
")
We will make the inference first by finding a maximum a posterior estimate for the hyperparameters via gradient based optimization. After this we will use grid integration and Markov chain Monte Carlo sampling to integrate over the hyperparameters.
# Set the options for the scaled conjugate optimization
o_source(text="
opt=optimset('TolFun',1e-3,'TolX',1e-3,'Display','iter');
")
# Optimize with the scaled conjugate gradient method
o_source(text="
gp=gp_optim(gp,x,y,'optimf',@fminscg,'opt',opt);
")
Make predictions of the underlying function on a dense grid and plot it. Below Eft_map is the predictive mean and Varf_map the predictive variance.
o_source(text="
[xt1,xt2]=meshgrid(-1.8:0.1:1.8,-1.8:0.1:1.8);
xt=[xt1(:) xt2(:)];
[Eft_map, Varft_map] = gp_pred(gp, x, y, xt);
figure(1)
clf
mesh(xt1, xt2, reshape(Eft_map,37,37));
hold on
plot3(x(:,1), x(:,2), y, '*')
axis on;
title('The predicted underlying function and the data points (MAP solution)');
drawnow;
")
We can also integrate over the hyperparameters. Following line performs grid integration and makes predictions for xt
o_source(text=" [gp_array, P_TH, th, Eft_ia, Varft_ia, fx_ia, x_ia] = gp_ia(gp, x, y, xt, 'int_method', 'grid'); ")
Alternatively we can use MCMC to sample the hyperparameters. (see gp_mc for details) The hyperparameters are sampled using default method, that is, slice sampling.
# Do the sampling (this takes approximately 5-10 minutes) # 'rfull' will contain a record structure with all the sampls # 'g' will contain a GP structure at the current state of the sampler o_source(text=" [gp_rec,g,opt] = gp_mc(gp, x, y, 'nsamples', 220); ") # After sampling we delete the burn-in and thin the sample chain o_source(text=" gp_rec = thin(gp_rec, 21, 2); ") # Make the predictions o_source(text=" [Eft_mc, Varft_mc] = gp_pred(gp_rec, x, y, xt); ")